Many diseases are determined by differences in yet undiscovered gene regulation. Thanks to plasmids and ‘barcodes’, the SuRE assay can map this relatively easily and extensively for the entire human genome.
‘The breakthrough came from the idea of using barcodes,’ says Bas van Steensel, professor of chromosome biology at the Erasmus University Medical Center and working at the Netherlands Cancer Institute, the research institute affiliated with the Antoni van Leeuwenhoek Hospital. ‘By attaching a random piece of sequence to each DNA fragment to be measured, we were able to track and identify it. This allowed us to scale up the entire process and we were able to map all the regulatory elements of the human genome in detail in one go.’

‘With SuRE we can map all regulatory elements of the human genome in detail in one go.’
Bas van Steensel
The development of this so-called Survey of Regulatory Elements (SuRE) by Joris van Arensbergen in the Van Steensel lab led to the establishment of the start-up Gen-X in 2017. The applications initially focused on gene therapy, but it soon became clear that much more could be done with the assay. And so, last year, the start-up Annogen was added, which focuses on cell therapy, drug development and agricultural biotechnology. What is the success story behind this technique?
Promoters and enhancers
All kinds of regulatory elements in the non-coding part of our DNA regulate the activity of genes (see box ‘ No junk DNA ‘). When a mutation in such a regulator causes disrupted gene expression, a disease can occur. But identifying and locating these mutations is incredibly difficult. With the current technique of genome-wide association studies (GWAS) you only get there partly, says Van Steensel. ‘This type of research gives poor resolution. We can say roughly in which part of the genome the mutation is located, but not exactly where. And that is necessary if you want to understand the disease and ultimately treat it better.’
The SuRE technique looks at the problem from the other side. ‘We first chop up the genome into millions of pieces of about five hundred base pairs each’, explains Joris van Arensbergen, CEO and founder of Gen-X and Annogen. ‘We then build a plasmid library from that. A standard plasmid contains a gene, but also a promoter to bring the gene to transcription. In our assay, that promoter is replaced by such an unknown piece of genome. We also add a barcode: a known sequence that we associate with that DNA element.’
The plasmids are then transfected into a cell line. ‘Genome pieces containing regulatory elements such as promoters and enhancers will then express the barcode’, says Van Arensbergen. ‘Using high-throughput sequencing, we then count each barcode in the RNA, and in this way we can read out the promoter and enhancer activity for the entire genome from which the library is made. And by then comparing different genomes, you will see which mutations influence regulator activity.’
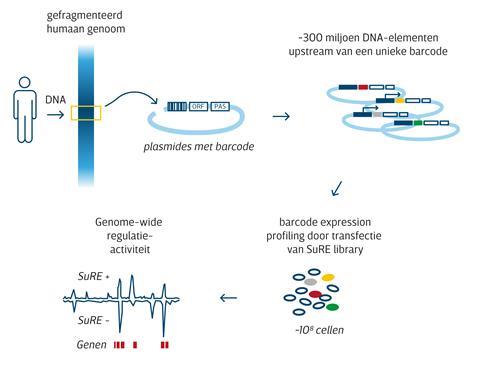
The technology behind SuRE
Genetic passport
There are many applications. In non-coding DNA, clues can often be found about the cause of diseases and disorders. One such disorder is autism. ‘Autism is largely hereditary’, says associate professor Raymond Poot (Erasmus MC). ‘But for more than 80% of people diagnosed with the disease, we still do not know exactly where the problem lies on the genome.’ This often involves mutations of one base pair, so-called single nucleotide polymorphisms (SNPs) in parts of the genome ranging from tens of thousands to more than one million base pairs long, so-called loci. ‘It is a mystery to us which SNPs in those loci are important.’
According to Poot, the only technique that can determine the important SNPs in such large parts of the genome is the SuRE technology. And so Poot will soon start a project together with Van Steensel and Hieab Adams, clinical geneticist and researcher at the Erasmus MC, to use this assay to map the gene regulation that is associated with an increased risk of autism. ‘We want to find those hidden mutations in regulatory elements’, Poot explains. ‘We do this by introducing a SuRE library into human neural stem cells and seeing where transcription takes place. These neural stem cells create the brain early in pregnancy. With this, we hope to better understand the autistic brain.’
‘Using SuRE in neural stem cells, we hope to better understand the autistic brain’
Raymond Poot
The ultimate dream is to eventually get such a high resolution that a ‘genetic passport’ of autism can be created. ‘On the basis of that, we can then determine whether someone has a greatly increased chance of this disorder’, says Poot. ‘This could even be done shortly after birth, for example in the heel prick screening. The earlier you determine an increased chance of autism, the sooner he or she can benefit from behavioural therapy.’
‘Van Steensel is thinking of a completely different application of SuRE: cancer research. ‘Almost all current publications in this field are about coding mutations. But there are also a huge number of mutations in the non-coding part of the genome. We want to tap into this black box with SuRE.’
Drug screenings
Gene regulation can be studied, but also used as a tool. For example, we are talking about cell therapy. ‘We do screenings for biotech companies that want to better control their process,’ says Van Arensbergen. ‘That then produces regulatory elements that can regulate or boost the expression of certain genes.’ A similar idea applies to gene therapy, which is done via start-up Gen-X. Van Arensbergen: ‘For greater efficiency but also safety, you can then look for promoters or enhancers that express the therapeutic gene highly, or precisely under certain conditions. For diseases in which inflammation plays a role, you look for an inflammation-inducible promoter, for example.’
A major advantage of SuRE is that you can generate a regulatory profile in any transfectable cell type based on DNA alone. Since you can now also easily order synthetic DNA, you actually only need the DNA sequence to then generate a SuRE library and a regulatory profile. ‘We are now looking at possible applications for DNA that is difficult to obtain, such as from the mammoth or the Neanderthal’, says Joris van Arensbergen, CEO of Annogen. ‘You then obviously have to transfect such libraries into comparable species, such as the elephant and human. We do not see a very large market for this, but we think it would be fun to participate in these kinds of projects.’

Joris van Arensbergen: ‘We conduct screenings for biotechnological companies, for example, that would like to better manage their processes.’
SuRE can also play a role in drug development. Currently, so-called drug screenings for toxicity are sometimes too limited. ‘For example, only one gene of one signal transduction pathway is examined’, Van Arensbergen explains. ‘As a result, you can miss off-target effects. Using the SuRE assay, we can find multiple regulatory DNA elements that serve as sensitive sensors to monitor multiple pathways.’
Another application in which Annogen recruits its customers is the production of recombinant proteins, for example as a medicine. ‘We can particularly help with very specific cell lines, such as algae. With SuRE we can then help increase production by helping to choose the right combination of promoter and enhancer for the expression of the recombinant gene.’
Gene editing
As beautiful as the technique is, according to Van Arensbergen it will have to be further developed. ‘We want to scale it up further to make it applicable for tens or hundreds of genomes. In addition, we have to reduce background expression even further in some systems. This initially occurred in plants, for example. We are also making the technique available for use in in vivo screens. For this we are continuously working on adjustments and variants of the SuRE vector.’
And perhaps there is another application in the offing: gene editing. With the advent of these technologies, instead of adding a complete therapeutic gene (as in gene therapy), you can specifically make small changes to the genome. ‘You can think of repairing a mutation in a gene,’ says Van Arenbergen. ‘But in this era of personalized medicine, gene editing of the non-coding part of the genome is also increasingly being looked at and we hope to make an important contribution here by testing thousands of potential edits with SuRE. That way, you only have to test the best ones in a model system.’
No ‘junk DNA’
Molecular geneticists are increasingly aware of the importance of the function of non-coding DNA. There are 20,000 to 25,000 genes contained in the three billion base pairs that make up the entire human genome – all genetic information contained in DNA – but all these genes make up only 1% of the total amount of DNA. So 99% of the genome has a non-coding function. The old term ‘junk DNA’ is way off the mark, as we now know: the non-coding part contains numerous regulatory elements that determine the level of expression of the genes. These have names such as promoters, enhancers, silencing elements and transcription binding sites. It is now clear that no less than 95% of all human characteristics and diseases have their origins in the non-coding part of the genome.






